Wings for NONMEM on the NESI Mahuika
Cluster
Home | Installation | Control Streams | Bootstrap
| Randomization Test | Visual
Predictive Check | Autocovariate | Files | References
Last Updated: 26 June 2025
This
web page describes the use of the NeSI Mahuika computing resource. Mahuika is
no longer available. It has been replaced by the HPC3 computing resource (see https://wfn.sourceforge.net/wfnhpc.htm).
This page is maintained only for historical reasons.
Prerequisites
You will need to have installed
several tools on the computer you will be using to run RJM tools before you can
do anything useful.
This is command line tool for starting a variety of tools for using
NONMEM.
2.
Mobaxterm
An X-windows terminal based GUI for access and use of NeSI server such
as Mahuika.
3.
Google
Authenticator or Authy installed
on your smartphone (Google or Authy) or web browser (Authy).
A 2 factor authentication tool required each time you use Mobaxterm to
access Mahuika.
4.
Gpg4win
An encryption tool used by Remote Job Management to use 2 factor
authentication.
5.
Cognex QR barcode reader
app installed on your smartphone.
Look for it in Google Play or Apple Store. Be sure to install the QR
reader with the yellow icon. It is used to extract the secret code required by
rjm_configure to set up a Windows computer to use Remote Job Management tools. The secret code may be shown by the NeSI site avoiding
the need to get it from the Cognex QR reader.
6.
References to “nesiname”
are the user name recognized by NeSI. For UoA users this is the same as the
username or UID.
Using Mobaxterm to
Maintain your NeSI Account
1.
You will need to use
Mobaxterm to setup your NeSI password. This part of the process is complex and
can be frustrating. Use the link below to make sure you have a NeSI login.
https://support.nesi.org.nz/hc/en-gb/sections/360000034315-Accessing-the-HPCs
Subsequently you can use
Mobaxterm to find, view and manage your files on Mahuika.
2.
Install Mobaxterm.
3.
Open Mobaxterm
4.
Set up a lander Session by
clicking on Session on the Mobaxterm main menu and then SSH.

5.
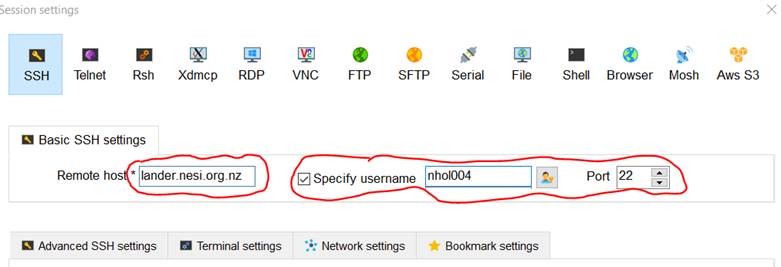
Set the Remote Host to
lander.nesi.org.nz. Use your NeSI username as default username with port 22.
6.
Click on the lander
session on the Mobaxterm home page to login to the login node. You will need to
do this in order to setup your 12 character NeSI password. Please open the link
below in a separate tab on your browser so you can refer to it easily while
setting up your password using Mobaxterm. Follow the instructions carefully one
by one.
https://support.nesi.org.nz/hc/en-gb/articles/360000335995-Setting-Up-Your-Password
7.
After you have setup your
NeSI password you should set up a session to access Mahuika.
NeSI Two Factor
Authentication
8.
When you register a
username and password with NeSI you will have to set up two factor
authentication. During the process you will be sent a web page with a QR code
image. BE SURE TO SAVE A COPY OF THE QR CODE IMAGE. Use the QR code image with
an authenticator app such as Authy or Google Authenticator to set up two factor
authentication for your NeSI account.
9.
This step may no
longer be necessary (2021-05-01). You will need the
QR code image to extract the QR secret code. The QR secret code is used by
automate the two factor authentication required to use NeSI (see below). To
extract the QR secret code you need to use the Cognex barcode scanner app. Go
to your smartphone app store e.g. Apple Store, search for Cognex, download and
install the app (yellow icon). Open the Cognex app and point your phone at the
QR code image. Once the image is captured you will have the choice to copy or
share the QR secret code. It is a long string of numbers that you will need to
copy later. So copy and save it in a text file so that you can find it easily.
Also email it to yourself (click on Share) in case you cannot locate your text
file.
Using Mobaxterm to Access
Mahuika
10. Open Mobaxterm
11.
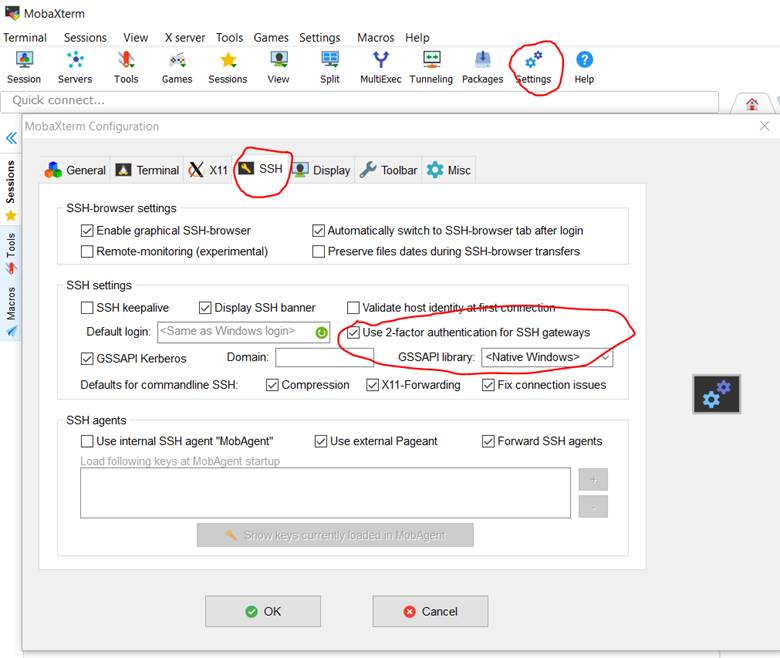
Enable 2 factor
authentication from main Mobaxterm menu by clicking on Settings then SSH. Be
sure to do this before you try to set up the Mahuika session.
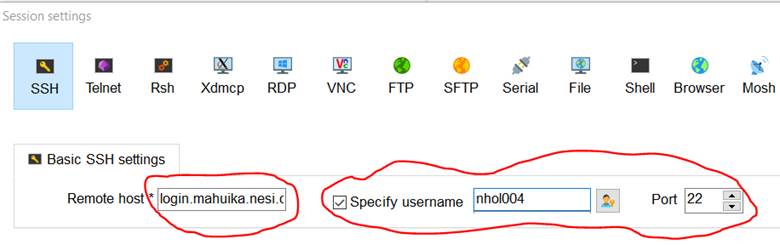
12. Set up a login.mahuika Session using SSH.
13. Set the Remote Host to login.mahuika.nesi.org.nz. Use your NeSI username
as default username with port 22.
14. Change Advanced SSH setting to use SSH-browser type of SCP (enhanced
speed)
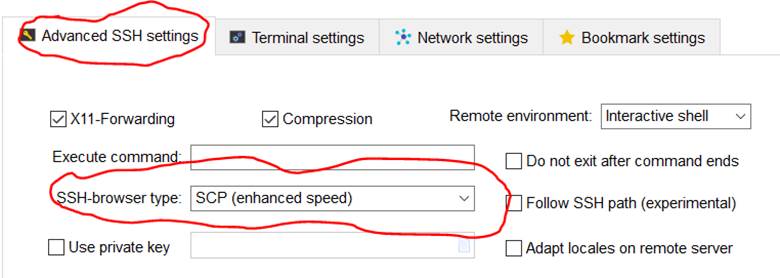
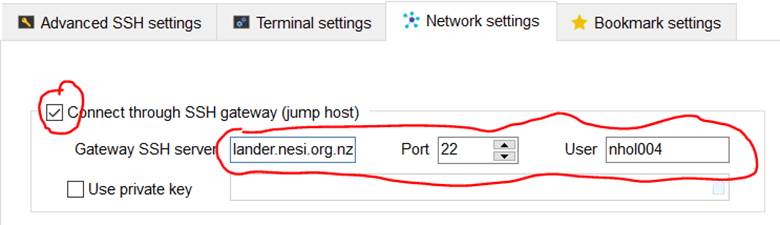
15. Change Network settings to login through gateway SSH server
lander.nesi.org.nz.
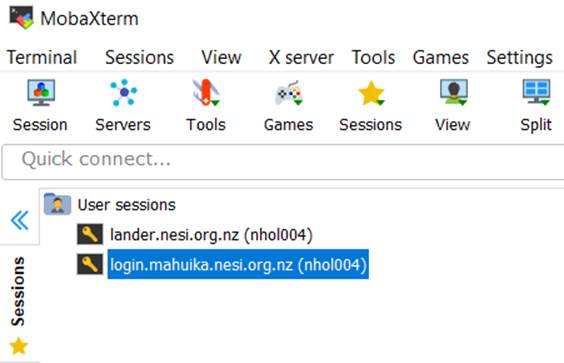
16. Click on the login.mahuika Session on the Mobaxterm home page to login
to Mahuika.
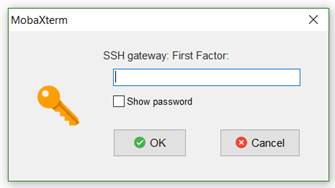
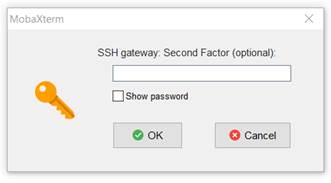
17. You will prompted by Mobaxterm for your first factor (your 12 character
NeSI password) then prompted for a second factor (the two factor authentication
code you obtain using Authy or Google authenticator). If this is successful you
will prompted again for the second factor code in the Mobaxterm terminal
window. DO NOT ENTER THE SECOND FACTOR CODE AGAIN. Just press Enter and you
will be logged into Mahuika.
Using Mobaxterm to See
your NeSI Home and Project Home Directories
18. When you are logged in to Mahuika you will be in your NeSI home
directory. Directories and files created using RJM tools are in on a different
file system (“nobackup”). You can use the left hand pane of Mobaxterm to view
your home directory.
19. You will need to be able to switch to the “nobackup” file system to view
your NeSI project home. In the right hand terminal window paste the following.
This creates a link to the project folder which you can use to quickly change
to the project home directory. Change uoa00106 to your own project
number as needed.
ln -s /nesi/nobackup/uoa00106 ~/MY_PROJECT
Your NeSI home directory
should look something like this after creating the MY_PROJECT link.
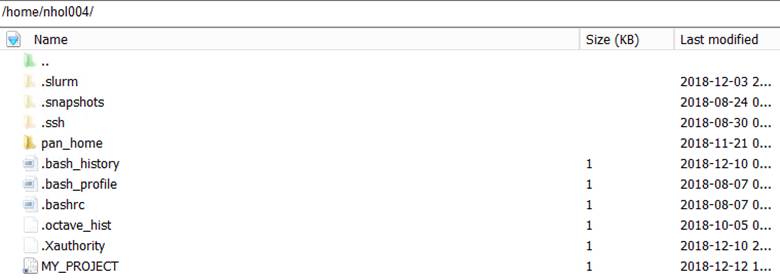
20. Click on MY_PROJECT. This will change the directory path in the left
hand window to the project directory on the “nobackup” file system.
21. In order to be simply return to your NeSI home directory you should
paste the following to the right hand terminal window. This creates a link to
your home directory to quickly go home. Change nesiname
to your NeSI user name as needed.
ln -s /home/nesiname/ MY_HOME
22. Double click on MY_HOME in the directory explorer to return to your NeSI
home directory.
23. Double click on MY_PROJECT in the directory window to go to the project
home directory.
24. Then double click on the directory named with your UPI. You will then
see a directory called rjm-jobs.
25. If you double click on rjm-jobs you can explore your job directories.
26. You can clean up all your files by selecting rjm-jobs then deleting it
(right click and delete or click on X delete icon). ONLY DO THIS WHEN YOU ARE
SURE ALL JOBS HAVE FINISHED RUNNING.
Advanced RJM Tools Options
27. The environment variable rjm_dir may be set to point to a different
directory for running jobs on the cluster e.g.
set rjmdir=/projects/myProject/myUPI/rjm_PK
You should change
myProject to your project and myUPI to your personal identifier. This can be
useful if you are running batches of jobs that you want to be able to easily
identify.
Installation to run WFN
with RJM tools on Mahuika
28. Install WFN version 751 (or later) and check that it works with nmgo theopd in the %WFNHOME%\run directory.
WFN 750 and earlier versions work with the Gpg4win version of RJM tools. This
method is not longer supported. The funcX and Globus version of RJM tools
should be used to access the NeSI Mahuika cluster
29. Jobs are submitted to the NeSI Mahuika cluster and results downloaded
using a set of Remote Job Management Tools.
30. Download the Remote Job Management tools zip archive. This version use
funcX and Globus:
31. Extract the files into your %WFNHOME%\bin directory.
Configuring RJM Tools
(funcX and Globus version)
32.
Read
the RJM documentation provided at:
https://chrisdjscott.github.io/RemoteJobManager/getting_started_nesi.html
33.
Run
rjm_nesi_update
with the --config and -ll debug options
rjm_nesi_update --config -ll debug
34.
This
will set up funcX and Globus. Note that this uses your default computer web browser
and does not work with older browsers like Microsoft Internet Explorer. You
should set your default web browser to a more modern browser such as Chrome.
35.
Run
rjm_confgure
36.
This
will ask for configuration information provided by rjm_nese_update to complete
the configuration on the specific computer where you run WFN.
Configuring RJM Tools
(Gpg4win version – no longer supported)
37. Install Gpg4win with the default options. This is used
to encrypt the QR secret code used as part of the 2 factor authentication
process.
38. Open a WFN command window.
39. Type the command rjm_configure, press enter and follow
the instructions to enter your NeSI login name and password. This needs to be
done only once by each user of a particular machine. The configuration dialog
is shown below. Your should enter your NeSI username where you see
<nesiname> and your project code, where you see <nesiproject>. The
project code is case sensitive e.g. enter uoa00106 not UOA00106.
40. You will need to enter the QR secret code. Open the
text file where you have saved the QR secret code and copy it to your
clipboard. When prompted by rjm_configure for the QR code secret then right
click once to paste it.
41. University of Auckland Pharmacometrics users have the
NeSI Project code uoa00106. Respond with ‘y’ to use the defaults when prompted
for file names.
>rjm_configure
Creating
configuration file C:\Users\<windowsname>\.remote_jobs\config.ini. Need
some information.
############################################################################################
Your NeSI username:
<nesiname>
NeSI password:
************
Repeat password:
************
QR code secret:
********************************************************
Default project code:
<nesiproject>
Use default values
for the other configuration parameters?
(Type y or Enter for
yes, or any other key for no) [y]?
Setting up password
store (this may take a few seconds)
#######################################################
gpg: checking the
trustdb
gpg: marginals
needed: 3 completes needed: 1 trust model: pgp
gpg: depth: 0 valid:
1 signed: 0
trust: 0-, 0q, 0n, 0m, 0f, 1u
gpg: next trustdb
check due at 2028-12-09
Done
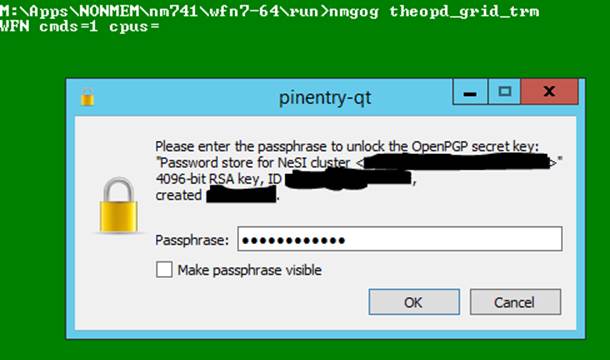
42. If you have not run rjm_configure properly then you may see something like this when you try to use
nmgog. You must try to run rjm_configure again.
> nmgog
theopd_grid_trm.ctl
WFN cmds=1 cpus=
Traceback (most
recent call last):
File "rjm_authenticate.py", line 8,
in <module>
File "site-packages\cer-0.1-py3.6.egg\cer\client\pypass\passwordstore.py",
line 52, in __init__
Exception: could not
find .gpg-id file
[112] Failed to
execute script rjm_authenticate
Remote Job Management
credentials are not valid for this run
43. Details are
stored in %USERPROFILE%\remote_jobs\config.ini
Some older
installations may still have an out of date value for the lander_host value. If
you are getting a connection actively refused error message then edit
config.ini so that lander_host is set to lander.nesi.org.nz
[CLUSTER]
lander_host=lander.nesi.org.nz
Using WFN with NeSI
44. Open a WFN window. You should now be able to use NeSI by calling nmgog:
nmgog theopd
45. RJM Tools Gpg42in version: The first time you use WFN with NeSI you will
be asked for a passphrase. The passphrase is the same as your NeSI password. It
will be remembered for subsequent runs until you logout (or restart your
computer).
46. The nmgog command will start
the job on the Mahuika cluster. When it finishes you should see the usual
results that are displayed by WFN.
47. The nmgog, nmbsg, nmbsig, nmrtg and nmgosimg commands work similarly to nmgo,nmbs,
nmbsi, nmrt and nmgosim but
submit NONMEM runs to the cluster. The number of cpus is set by default to 4
and walltime to 4:0:0 (4 hours) for cluster jobs. These defaults can be changed
by setting the cpus and walltime environment variables before the WFN commands.
set CPUS=24
Note that if you ask for a lot of CPUs your job may be
put into a wait queue until there are enough CPUs available.
The WALLTIME variable is specified in the
format hh:mm:ss. It controls the total run time for your job. You might
estimate this from a run on a typical Windows machine and divide by 2 (it
should be at least 2 times faster with 4 CPUs). The default time for checking that
the job is finished is 10 seconds. You may set the BATCHWAIT variable to
a more suitable time if you have long jobs.
Request up to 24 hours for
job to run
set WALLTIME=24:00:00
Check every 60 seconds to see if jobs have finished
set BATCHWAIT=60
48. The default memory requested is 250 megabytes. If a run fails then check
stderr.txt in the WFN results folder. This may indicate not enough memory. Try
increasing the memory request in steps of 250M. An error such as “compiler
failure” also suggests increasing the memory.
The memory request can be
changed with the NMMEM environment
variable. Note that memory size must be specified as an integer with M or G
suffix.
Request 750 megabytes of
memory:
set NMMEM=750M
Request 1 Gigabyte of
memory:
set NMMEM=1G
Limitations and Special
Cases
49. Because of the way two factor authentication works on Mahuika there are
2 known conflicts with RJM tools Gpg4win version. These can be overcome by:
i) Opening Mobaxterm before you try and use rjm_tools.
ii) Waiting for each rjm_batch_submit to complete the submission
process before trying to initiate a new rjm_batch_submit job. This means
typically waiting about 30 seconds but it will depend on the complexity of the
files you need to set up and transfer with each rjm_batch_submit task.
50. Gpg4win version: The environment variable nmwaitonly may be set to y eg.
set nmwaitonly=y
This is rarely needed but it is possible that jobs
have run and completed but have not yet been downloaded (e.g. you logged out or
restarted your computer). By setting nmwaitonly to y and rerunning the cluster
command (e.g. nmbsg) then the
rjm_batch_wait tool will be restarted, no new job will be submitted and the
results will be downloaded (after some delay). Use of nmwaitonly is no longer
required when using the RJM tools version using funcX and Globus.
51. Remember to unset nmwaitonly or set it to n in order to restore the
default behavior.
52. WFN users may occasionally want to use rjm_batch_cancel. Note that this
command is executed from the WFN command window in the directory where you
started a cluster job or batch of jobs. It uses the *localdirs.txt file created
by WFN. If this file is missing or if the directories listed in this file are
missing then this rjm tool command will not work. Individual jobs can be
cancelled using the rm_batch_cancel command e.g.
rjm_batch_cancel -f theopd_localdirs.txt -z 10
This command would be used
to stop a job started with a WFN run name of theopd.
53. Individual jobs can also be cancelled in the Mobaxterm command window
using the scancel command. This
requires that you know the job id. Using sacct
(see below) shows job ids.
scancel jobid
54. nmstopg will cancel all jobs started
by rjm_batch_submit (e.g. nmgog, nmbsg).
nmstopg
55. Memory usage for jobs on a particular date can be obtained using this
command at the login node command prompt (use Mobaxterm) with a suitable user
id and date:
sacct -u nesiname -S
2018-12-10
or if you have a jobid (eg. From using squeue while
the job is running) this will show details of a running or completed job. The
completed job stats show the maximum memory usage (maxRSS). This may be useful
in estimating the requested memory size (see nmmem).
sacct -j jobid
56. Job status can be obtained using
this command at the login node command prompt (use Mobaxterm) with a suitable
user id:
squeue -u nesiname
The squeue command shows job numbers which can be used
with sview. The sview command can be used to find information about each job
but is rather clumsy to use when trying to find a particular job.
sview
57. From mid-February 2021 WFN maintains a log of jobs run using NeSI. WFN
includes a smrg command that retrieves job statistics derived from the job
numbers and merges the output of nm_seff and sacct to show CPU, memory and
walltime efficiency.
smrg
The results of smrg are
collected into a file called smrg_stat.csv. This can be viewed using Excel and
specific rows selected using the Excel data filter. Each set of job statistics
starts with the date and time of the run and the job ID (slurm job number).
This example shows 3 rows
selected with Date containing ’04-06’. The results for each job include the
user name, the state of the job when it finished, and 4 sections with
statistics describing efficiency of requested CPU, memory and walltime.
The CPU section shows the number
of CPUs requested and the CPU efficiency. When CPUeff% is less than 2% it
typically means the job finished with an error detected by NM-TRAN or NONMEM.
Models similar to Job number 18921115 which ran successfully had a CPUeff% of
99%. For reasons currently unexplained jobs running with NM7.5.0 report a State
of ‘FAILED’ even though the NONMEM job completed normally. The theopd test job
ran successfully but with low CPU efficiency because each individual has only a
few observations.
|
Date |
Job ID |
User |
State |
CPUS |
CPUeff% |
|
2021-04-06_17.51.03.219209 |
18921115 |
jmor616 |
COMPLETED |
24 |
1.25 |
|
2021-04-06_08.22.38.430877 |
18924529 |
nhol004 |
FAILED |
4 |
16.67 |
|
2021-04-06_08.44.33.746999 |
18925119 |
nhol004 |
COMPLETED |
1 |
29.58 |
The memory section shows the
requested memory (ReqMem). By default this is 250Mc for NONMEM jobs. The ‘c’
suffix indicates this is memory requested per core (similar to per CPU). The
user can request more memory by changing the NMMEM environment variable which
is also shown in this section. The main memory demand for NONMEM is during job
compilation and the memory required is similar for both small and large NONMEM
models. The Memory values shows the memory used by all the parallel tasks. This
increases in proportion to the number of CPUs. The MaxRSS statistic is hard to
interpret. One definition
is "Maximum individual resident set size out of the group of resident
set sizes associated with all tasks in job." When CPUeff% is high it is
usually several times bigger than Memory. It is not clear how it can be smaller
than Memory. The MEMeff% statistic is somehow related to ReqMem and Memory or
MaxRSS but it is not clear how.
|
Date |
Job ID |
ReqMem |
NMMEM |
Memory |
MaxRSS |
MEMeff% |
|
2021-04-06_17.51.03.219209 |
18921115 |
250Mc |
250M |
11.72GB |
24K |
0.16 |
|
2021-04-06_08.22.38.430877 |
18924529 |
250Mc |
250M |
1.95GB |
759K |
0.14 |
|
2021-04-06_08.44.33.746999 |
18925119 |
20Mc |
20M |
40.00MB |
94K |
15.94 |
The walltime section shows
the Elapsed time the job ran (d-hh:mm:ss), the TotalCPU time which is approximately
Elapsed time multiplied by the number of CPUs, and Walltime (the clock time
that elapses from starting to finishing the job). The WallEff% is the percent
of Walltime taken by Elapsed.
|
Date |
Job ID |
Elapsed |
TotalCPU |
Walltime |
WallEff% |
|
2021-04-06_17.51.03.219209 |
18921115 |
0:00:10 |
00:00.0 |
1-00:00:00 |
0.01 |
|
2021-04-06_08.22.38.430877 |
18924529 |
0:00:03 |
00:07.1 |
4:00:00 |
0.1 |
|
2021-04-06_08.44.33.746999 |
18925119 |
0:03:33 |
0:00:00 |
0:05:00 |
71 |
The final section indicates
the NONMEM version (NMVER) and the job name. For NONMEM jobs this will be the
same as the model file name with a suffix indicating the Windows shell command
counter for that job (‘_cmd1’). WFN cluster commands that run multiple jobs in
parallel such as nmbsg are filtered to show only the first shell command.
|
Date |
Job ID |
NMVER |
Name |
|
2021-04-06_17.51.03.219209 |
18921115 |
744 |
sevo_fixPKPD_popM_popP_popR_fixed_fixE0_d_cmd1 |
|
2021-04-06_08.22.38.430877 |
18924529 |
750 |
theopdg_cmd1 |
|
2021-04-06_08.44.33.746999 |
18925119 |
750 |
smrg_cmd1 |
The smrg command may be
run at any time. It takes just under 4 minutes to complete with a 2 month
collection of jobs. If someone else is running smrg you will get a warning
message and the previous most recent version of the smrg_stat.csv file will be
copied to the directory you used to call smrg.
58. Other commands to find out about jobs are described here:
https://support.nesi.org.nz/hc/en-gb/articles/360000205215-Useful-Slurm-Commands