Wings for NONMEM on the NeSI HPC3
Home | Installation | Control Streams | Bootstrap
| Randomization Test | Visual
Predictive Check | Autocovariate
| Files | References
Last Updated: 2 February 2026
Contents
Managing Files on HPC3 and Checking
on HPC3 job progress
Using NeSI ondemand to Access your
NeSI home directory
Using Slurm To Show The Status Of
Running Jobs
Keeping Track of HPC Job Files
How To Retrieve Results From A
Failed Job
Connection Used To Submit The Job
Has Stopped
History
An earlier
system for accessing NeSI computing was called the NeSI
grid.
HPC3 is
High Performance Computing System 3. On 1 July 2025, the roles, services
and technologies of New Zealand eScience Infrastructure (NeSI) were integrated
into the crown-owned company, Research and Education Advanced Network New
Zealand (REANNZ). The REANNZ logo appears on some output screens. NeSI is the
system that implements access and running jobs on HPC3.
Prerequisites
You will need to have installed WFN on
a Windows computer. WFN is a command line tool for using NONMEM.
Wings
for NONMEM (WFN) has links to the New Zealand eScience Infrastructure
(NeSI) system high performance computing resources (HPC3). When using WFN this
is primarily used to run NONMEM jobs using parallel processing on multiple CPUs (‘nodes’) on HPC3.
The link between WFN running on a
Windows computer and HPC3 is managed by the Remote Job Management (RJM) system.
RJM is a set of tools created by NeSI most recently maintained by Chris Scott.
The RJM tools have been primarily implemented to work with WFN. In order to use the RJM system you will need to run rjm_configure.
IMPORTANT
NeSI HPC3 is only accessible by applying for an
account and registration of individual users.
Apply for access with https://www.nesi.org.nz/researchers/apply-access-our-services
before trying to run rjm-configure. This only needs to be
done once for each user.
rjm-configure needs to be run on each computer you
plan to connect to HPC3. rjm-configure is located in
the wfn7\bin directory. You can run it by opening a WFN window and typing
rjm_configure
You will be prompted to provide a NeSI
user name (used to apply for access to NeSI) and a
NeSI project name (uoa00106 for Auckland Pharmacometrics Group users). rjm-configure
will suggest a directory name for storing NONMEM related files on HPC3. You
should accept the suggested name.
During the process of configuration you will be prompted to provide credentials
for access to NeSI. The images below show examples. You should accept the
defaults.
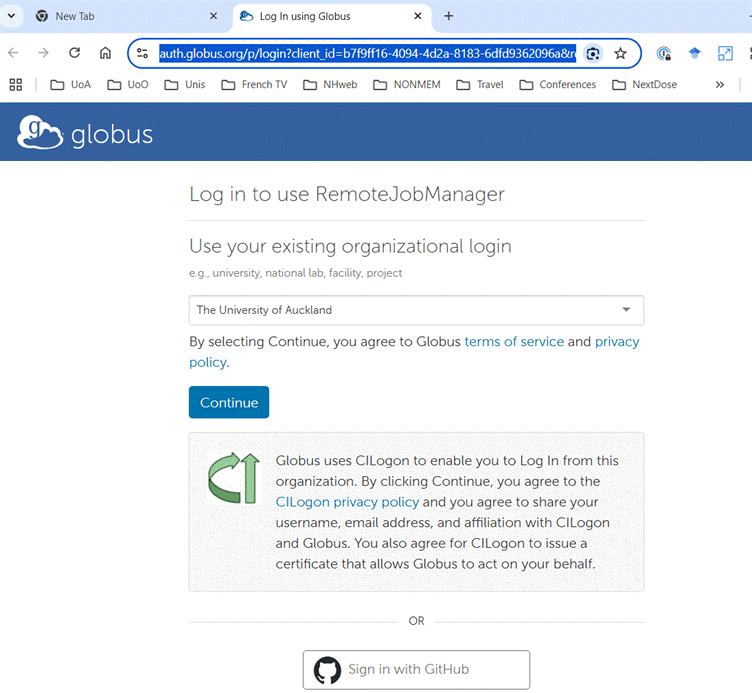
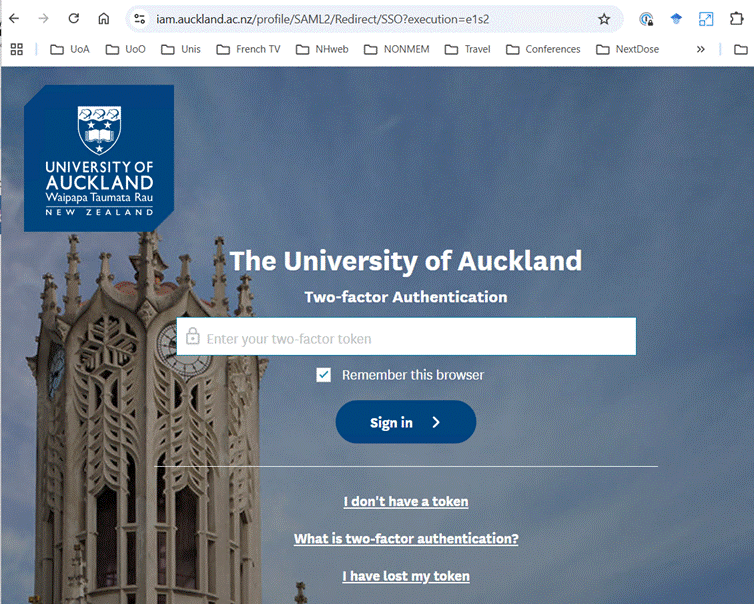
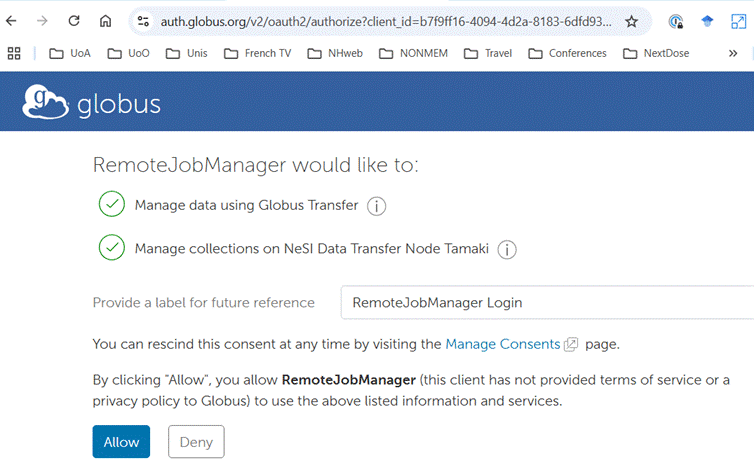
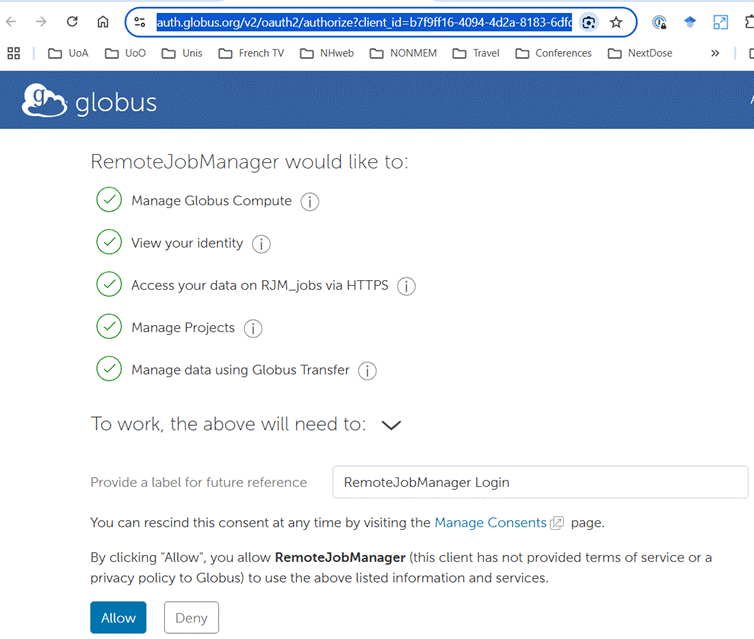
Once you
complete rjm-configure you can start using WFN to submit
NONMEM jobs to HPC3.
Managing Files on HPC3 and Checking on HPC3 job
progress
Using
NeSI ondemand to Access your NeSI home directory
Open NeSI ondemand
1.
https://ondemand.nesi.org.nz/public/
2.
Click on Login (you may be
prompted to show a 2 factor authentication code shown
on Google Authenticator).
3.
The ondemand system opens in the Dashboard page with the following options:
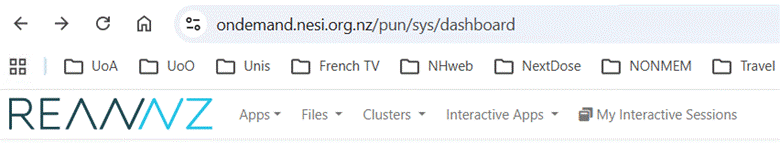
4.
Open a File manager window
by clicking on Files
then Home Directory so you can see the
directories and files stored in your NeSI home directory.
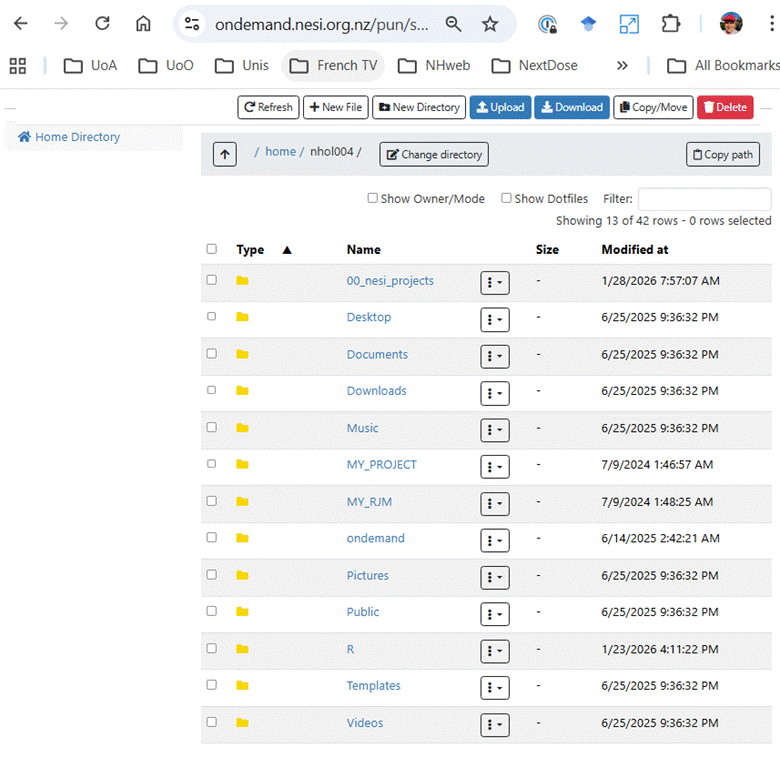
5.
Return to the dashBoard page (back-arrow). Open an HPC3 NeSI terminal
window by clicking on Clusters then _NeSI HPC Shell Access
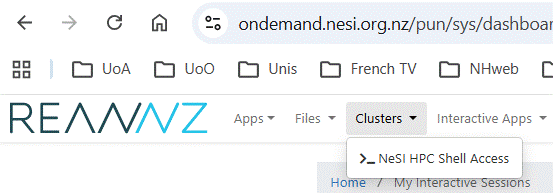
Or use this link:
https://ondemand.nesi.org.nz/pun/sys/shell/ssh/login.hpc.nesi.org.nz
This terminal window gives you command line access to
your files stored on the HPC3 system and access to the slurm operating system
commands.
![]()
Ask for a long directory
listing by typing the command ls -l
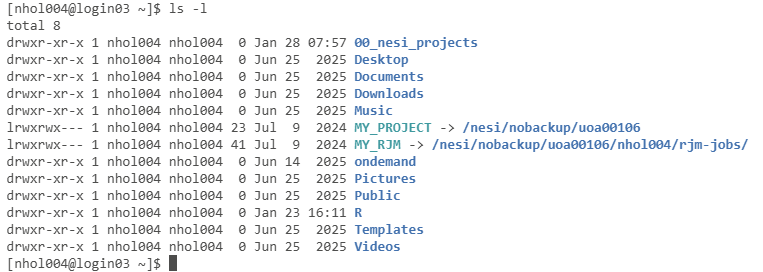
Setting
Up Symbolic Links
This is my NeSI home directory. There are two symbolic
links MY_PROJECT and MY_RJM that provide a simple way to access the uoa00106
project directory and your personal set of rjm-job files. It is recommended to
create these links by copying and pasting the following commands into the
terminal window. Be sure to change nesiname to
your NeSI user name
ln -s /NeSI/nobackup/uoa00106
~/MY_PROJECT
ln -s
/NeSI/nobackup/uoa00106/neSiname/rjm-jobs/ ~/MY_RJM
The ~ before the symbolic link name refers to your
NeSI home directory. This is the location where the symbolic link will be
created.
Using
Slurm To Show The Status Of
Running Jobs
Slurm is a specialized operating system used to manage
multiple users with access to the HPC3 system of compute nodes. Slurm may be
used to query how jobs are running on HPC3.
Use the squeue command to see the status
of running jobs e.g
squeue -A uoa00106
The -A uoa00106 option shows all jobs associated with
members of our NeSI group (uoa00106)
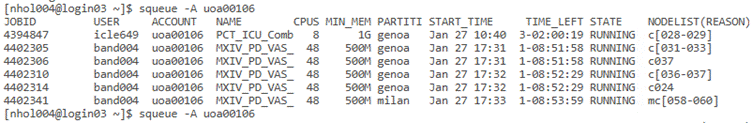
The -u nesiname option shows the jobs for a specific user e.g. my nesiname is nhol004.
squeue -u nhol004
![]()
The CPUS column in the output is currently two times
the number of CPUs requested when using WFN. This is because most of the actual
CPUs can perform a second set of tasks using hyperthreading. There are slurm
commands to enable hyperthreading but by default hyperthreading is not enabled.
Keeping
Track of HPC Job Files
Use the ondemand page dashboard
to open a session app. I prefer to use the VS Code (“Visual Studio”)
interactive app. Jupyter Lab offers similar functionality but some features
such as refreshing the file list do not seem to work. See the section below Ondemand Dashboard Apps
for more information. The following images show how to use the VS Code app to
keep track of HPC job files.
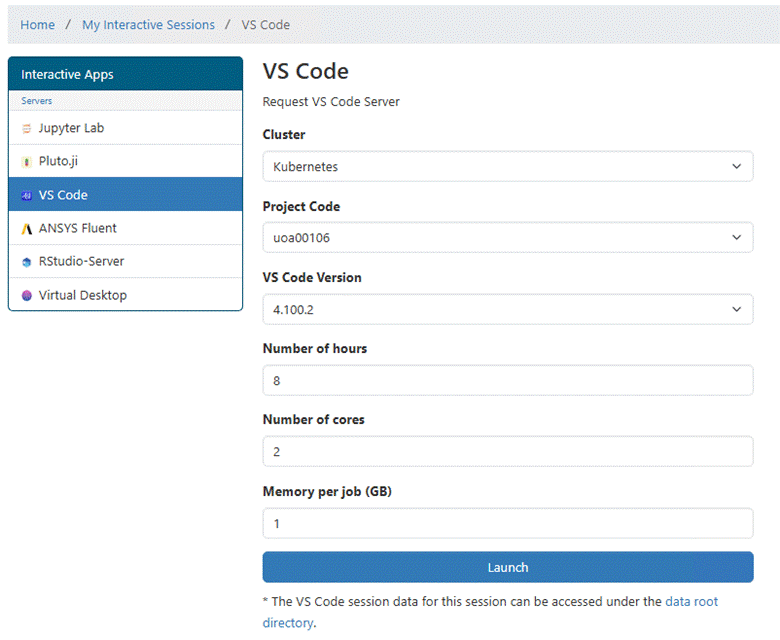
Clicking on VS Code will open the following window.
Currently the maximum number of hours is 8. That means you will need to open a
new VS Code app session after 8 hours.:
After launching the session app
you will see a window indicating the app is starting. This may take 10-30
seconds.
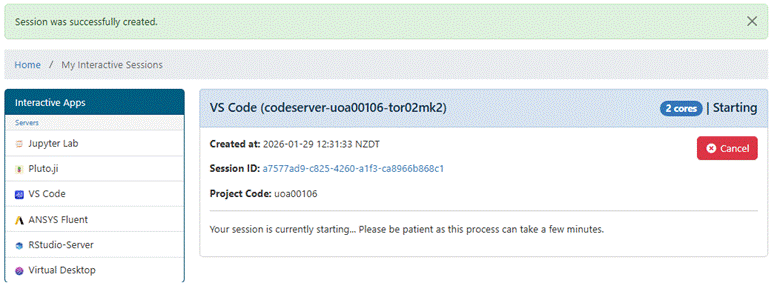
Once started, the
window shows the app is running.
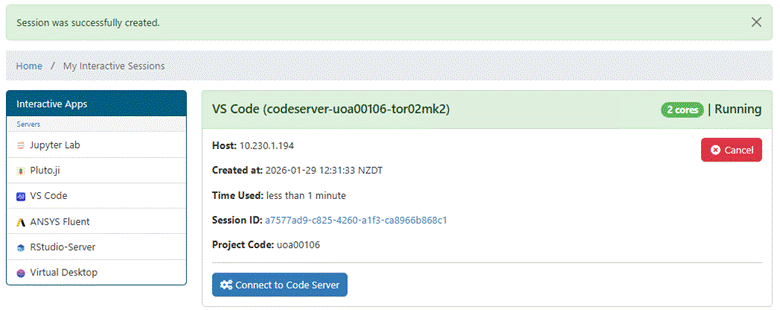
Use the Connect button to open the app. When the VS
Code app opens the upper right corner of the opening page looks like this:
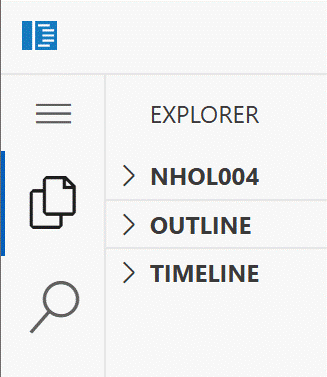
You may need to click on your user
name then the explorer symbol (overlapping pages) to open the user home
page.
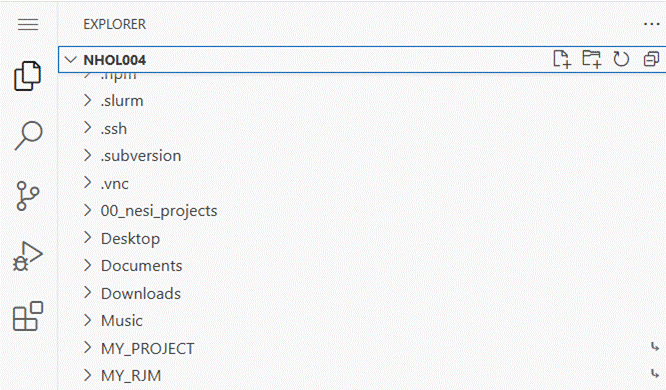
Scroll down to find a folder you are interested in
then click to open e.g. use MY_RJM to see the folders associated with recent
NONMEM runs. If the job has completed successfully the
folder contains an output.zip file containing results that are transferred to
WFN. If the job is still running then click on the job
folder name and look for the OUTPUT file.
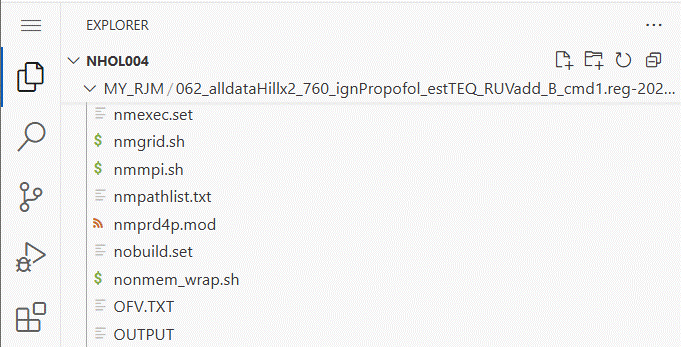
Click on OUTPUT and a window should open showing the OUTPUT file contents.
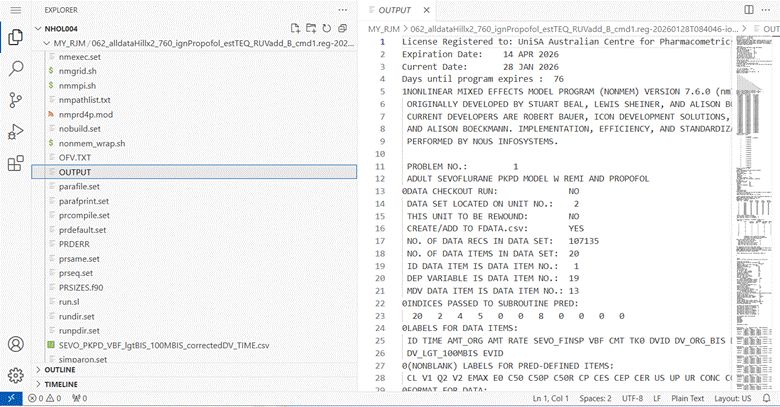
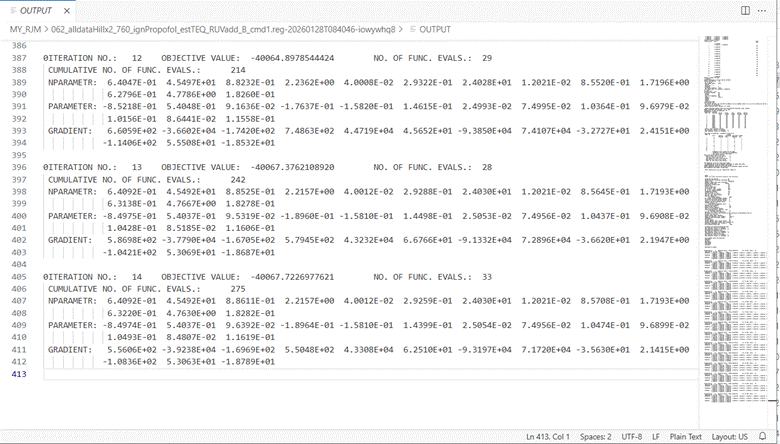
Click on the OUTPUT window and scroll down to the
bottom to find the results of the latest iteration. On the right
hand side of the window is a map of the lines in the file which can be
useful to find a section that interests you.
To refresh the EXPLORER window
hover your mouse over the file folder the click on revolving arrow symbol. This
refreshes the folder contents list and should also refresh the OUTPUT page.
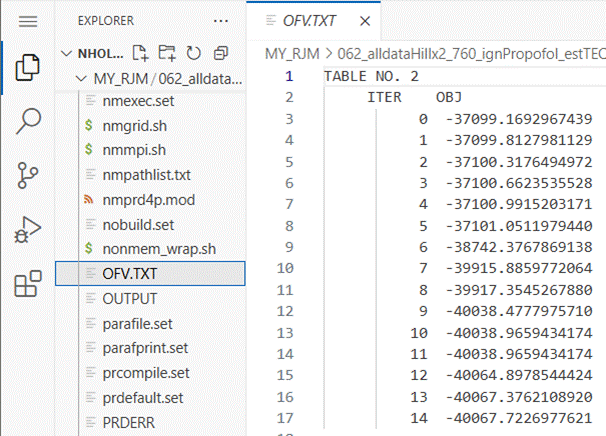
If you click on the OFV.TXT file name you can see a
list of objective function values (OFV).
When jobs have finished you
should consider deleting the run folder to release storage resources. You can
do this by opening a terminal window by clicking on the menu icon (3 bars) at
the upper right of the app window.
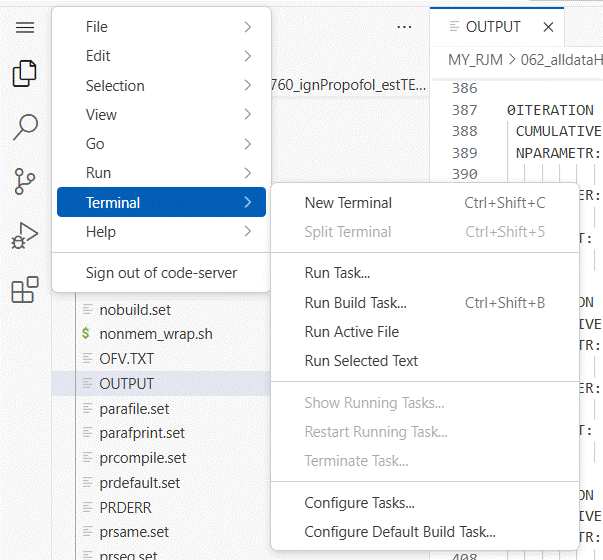
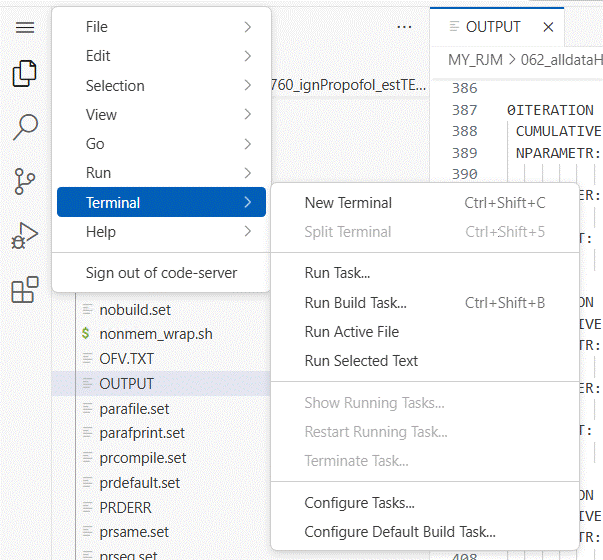
The terminal window opens in your NeSI home folder.
You can list the contents with the ls -l command typed into the
terminal window.
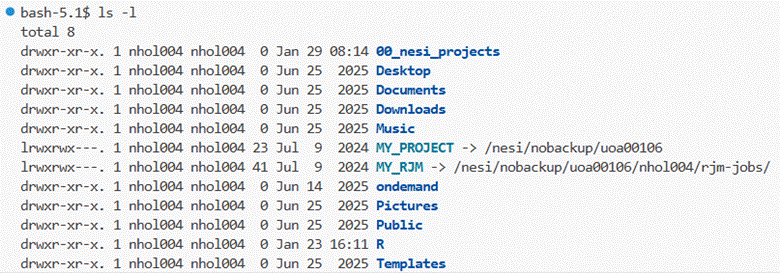
You will need to change directory to MY_RJM by typing
the cd MY_RJM command followed by ls-l to list the folders and files:

You can delete a folder and its contents with a
command such as:
rm -R 062*
Folders need to be removed recursively (rm -R)
But you should be cautious using the “*” wildcard because it will
delete all folders and files which start with “062”. This wildcard is useful
if you have 100 similar folders created by a bootstrap run with 100 replicates.
To delete just one folder it
is best to specify the full directory name. This can be done by double clicking
on the folder name which copies the name into the clipboard then type “rm -R “ followed by a right-click on the mouse which should paste
the full name after the partially typed command. Check that it looks correct
then press “Enter” to remove the folder.
It is not essential to delete folders
but it does help you to identify folders you are interested in and is “good
citizenship” not to consume resources once you have finished with them.
Eventually the folder will be deleted by NeSI based on their automated deletion
policy.
Once you have finished with the app you should return
to the ondemand dashboard session page, cancel the app then delete the saved
copy of the app in order release computing and storage resources.
Advanced
RJM Tools Options
The environment variable rjm_dir may be set to point to a different directory for running jobs on HPC3
e.g.
set rjmdir=/projects/myProject/myName/rjm_PK
You should change myProject
to your project and myName to your nesiname. This can be useful if you are running batches of jobs that you want to
be able to identify more easily.
Using
WFN with NeSI
1. Open a WFN window. You should now be able to use NeSI by calling nmgog:
nmgog theopd
2.
The nmgog command will start the job on HPC3. When it finishes you should
see the usual results that are displayed by WFN.
3.
The nmgog, nmbsg, nmbsig, nmrtg and nmgosimg commands
work similarly to nmgo,nmbs, nmbsi, nmrt and nmgosim but submit NONMEM runs to the cluster. The number of
cpus is set by default to 4 and walltime to 4:0:0 (4
hours) for cluster jobs. These defaults can be changed by setting the cpus and walltime environment variables before the WFN
commands.
set CPUS=24
Note that if you ask for a
lot of CPUs your job may be put into a wait queue until there are enough CPUs
available.
The WALLTIME
variable is specified in the format hh:mm:ss. It controls the total run time
for your job. You might estimate this from a run on a typical Windows machine
and divide by 2 (it should be at least 2 times faster
with 4 CPUs).
The default time for
checking that the job is finished is 10 seconds. You may set the BATCHWAIT
variable to a more suitable time if you have long jobs.
Request up to 24 hours for
job to run
set WALLTIME=24:00:00
Check every 60 seconds to see if jobs have finished
set BATCHWAIT=60
4.
The default memory
requested is 500 gigabytes. If a run fails then check stderr.txt in the WFN results folder. This may
indicate not enough memory. Try increasing the memory request in steps of 500
megabytes using the NMMEM
variable. An error such as “compiler failure” also suggests increasing the
memory.
The memory request can be
changed with the NMMEM environment
variable. Note that memory size must be specified as an integer with M or G
suffix.
Request 500 megabytes of
memory:
set NMMEM=500M
Request 2 gigabytes of
memory:
set NMMEM=2G
Using
nfmenv.bat
The Windows environment options such as CPUS and NMMEM
may need to be tailored to specific NONMEM projects. The nmenv.bat script is useful for specifying these
environment variables when opening a WFN window. The nmenv.bat should be placed in the directory where the WFN window opens. This is
typically the NONMEM directory used to store NM-TRAN control streams for the
project. The title command in nmenv.bat is useful for showing key environment
variables.
@echo off
rem nmenv.bat
set args7=
set CPUS=20
set NMMEM=2G
title Project CPUS=%CPUS%
The args7 variable is by default set to -prdefault in order to use pre-compiled PREDPP code. When
the NM-TRAN specifies changes to SIZES then the args7 variable should be unset. It is often useful to put this in nmenv.bat. Note that if you wish to run nmgo at the command line it is important to unset the CPUS variable. The CPUS variable should only be set when using WFN commands that use HPC3.
Slurm
Commands
1.
Memory usage for jobs on a
particular date can be obtained using this command at the login node command
prompt (use Mobaxterm) with a suitable user id and date:
sacct -u nesiname -S 2018-12-10
or if you have a jobid (eg.
From using squeue while the job is running) this will show details of a running
or completed job. The completed job stats show the maximum memory usage (maxRSS). This may be useful in estimating the requested
memory size (see nmmem).
sacct -j jobid
2.
Job status can be obtained using this command
at the login node command prompt with a suitable user id:
squeue -u nesiname
3.
The job status of all
users in the Auckland Pharmacometrics Group project can be displayed using:
squeue -A uoa00106
4.
The squeue command shows job numbers which can
be used with sview. The sview
command can be used to find information about each job but
is rather clumsy to use when trying to find a particular job.
sview
5.
From mid-February 2021 WFN
maintains a log of jobs run using NeSI. WFN includes a
smrg command that retrieves job statistics derived
from the job numbers and merges the output of nm_seff
and sacct to show CPU, memory and walltime
efficiency.
smrg
The results of smrg are collected into a file called smrg_stat.csv. This
can be viewed using Excel and specific rows selected using the Excel data
filter. Each set of job statistics starts with the date and time of the run and
the job ID (slurm job number).
This example shows 3 rows
selected with Date containing ’04-06’. The results for each job include the user name, the state of the job when it finished,
and 4 sections with statistics describing efficiency of requested CPU, memory
and walltime.
The CPU section shows the
number of CPUs requested and the CPU efficiency. When CPUeff%
is less than 2% it typically means the job finished with an error detected by
NM-TRAN or NONMEM. Models similar to Job number
18921115 which ran successfully had a CPUeff% of 99%.
For reasons currently unexplained jobs running with NM7.5.0 report a State of
‘FAILED’ even though the NONMEM job completed normally. The theopd test job ran
successfully but with low CPU efficiency because each
individual has only a few observations.
|
Date |
Job ID |
User |
State |
CPUS |
CPUeff% |
|
2021-04-06_17.51.03.219209 |
18921115 |
jmor616 |
COMPLETED |
24 |
1.25 |
|
2021-04-06_08.22.38.430877 |
18924529 |
nhol004 |
FAILED |
4 |
16.67 |
|
2021-04-06_08.44.33.746999 |
18925119 |
nhol004 |
COMPLETED |
1 |
29.58 |
The memory section shows
the requested memory (ReqMem). By default
this is 250Mc for NONMEM jobs. The ‘c’ suffix indicates this is memory
requested per core (similar to per CPU). The user can
request more memory by changing the NMMEM environment variable which is also
shown in this section. The main memory demand for NONMEM is during job
compilation and the memory required is similar for both small and large NONMEM
models. The Memory values shows the memory used by all the parallel tasks. This increases in proportion to
the number of CPUs. The MaxRSS statistic
is hard to interpret. One definition is "Maximum
individual resident set size out of the group of resident set sizes associated
with all tasks in job." When CPUeff% is high it
is usually several times bigger than Memory. It is not clear how it can be
smaller than Memory. The MEMeff% statistic is somehow
related to ReqMem and Memory or MaxRSS but it is not clear how.
|
Date |
Job ID |
ReqMem |
NMMEM |
Memory |
MaxRSS |
MEMeff% |
|
2021-04-06_17.51.03.219209 |
18921115 |
250Mc |
250M |
11.72GB |
24K |
0.16 |
|
2021-04-06_08.22.38.430877 |
18924529 |
250Mc |
250M |
1.95GB |
759K |
0.14 |
|
2021-04-06_08.44.33.746999 |
18925119 |
20Mc |
20M |
40.00MB |
94K |
15.94 |
The walltime section shows
the Elapsed time the job ran (d-hh:mm:ss),
the TotalCPU time which is approximately Elapsed time
multiplied by the number of CPUs, and Walltime (the clock time that elapses
from starting to finishing the job). The WallEff% is
the percent of Walltime taken by Elapsed.
|
Date |
Job ID |
Elapsed |
TotalCPU |
Walltime |
WallEff% |
|
2021-04-06_17.51.03.219209 |
18921115 |
0:00:10 |
00:00.0 |
1-00:00:00 |
0.01 |
|
2021-04-06_08.22.38.430877 |
18924529 |
0:00:03 |
00:07.1 |
4:00:00 |
0.1 |
|
2021-04-06_08.44.33.746999 |
18925119 |
0:03:33 |
0:00:00 |
0:05:00 |
71 |
The final section
indicates the NONMEM version (NMVER) and the job name. For NONMEM jobs this
will be the same as the model file name with a suffix indicating the Windows
shell command counter for that job (‘_cmd1’). WFN
cluster commands that run multiple jobs in parallel such as nmbsg are filtered
to show only the first shell command.
|
Date |
Job ID |
NMVER |
Name |
|
2021-04-06_17.51.03.219209 |
18921115 |
744 |
sevo_fixPKPD_popM_popP_popR_fixed_fixE0_d_cmd1 |
|
2021-04-06_08.22.38.430877 |
18924529 |
750 |
theopdg_cmd1 |
|
2021-04-06_08.44.33.746999 |
18925119 |
750 |
smrg_cmd1 |
The smrg
command may be run at any time. It takes just under 4 minutes to complete with a 2 month collection
of jobs. If someone else is running smrg you will get
a warning message and the previous most recent version of the smrg_stat.csv
file will be copied to the directory you used to call smrg.
6.
Other commands to find out
about jobs are described here:
https://support.NeSI.org.nz/hc/en-gb/articles/360000205215-Useful-Slurm-Commands
Ondemand
Dashboard Apps
The ondemand dashboard provides several apps that
might be useful. These apps require the user to
configure some settings for the app. Once launched the app takes a minute or so
to set itself up. Once it is running you can then click on Launch to get it
started. You can use the dashboard to cancel a running app so that it no longer
consumes HPC3 resources.
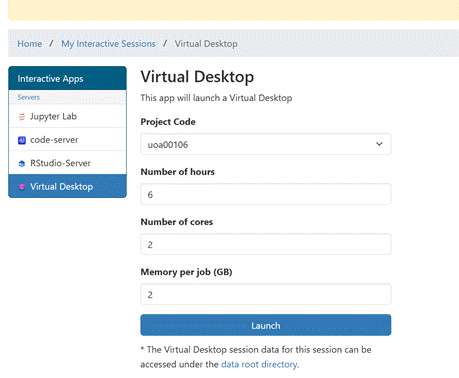
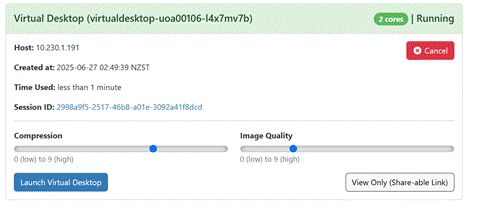
The Virtual Desktop looks
like this:
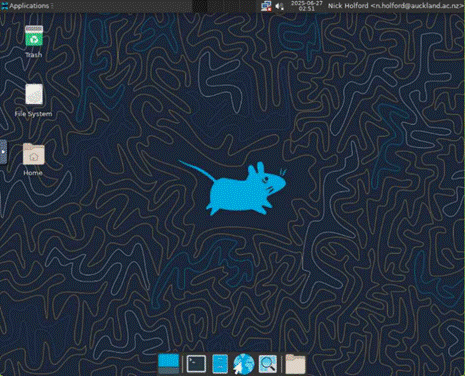
The Virtual Desktop offers
a File System view. By clicking on your NeSIname you
can see directories and files in your home directory. This is an alternative to
using the Files view offered in the ondemand dashboard.
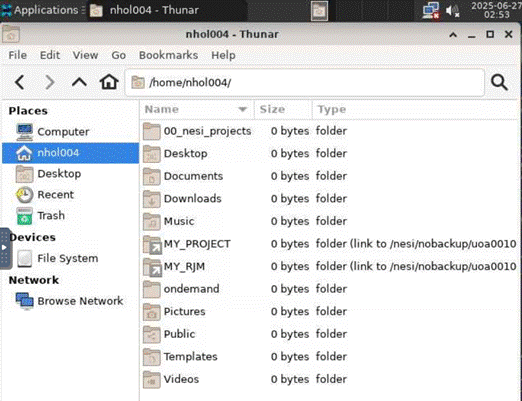
It also offers a terminal
view.
The Jupyter lab app offers
a combined File system and terminal view.
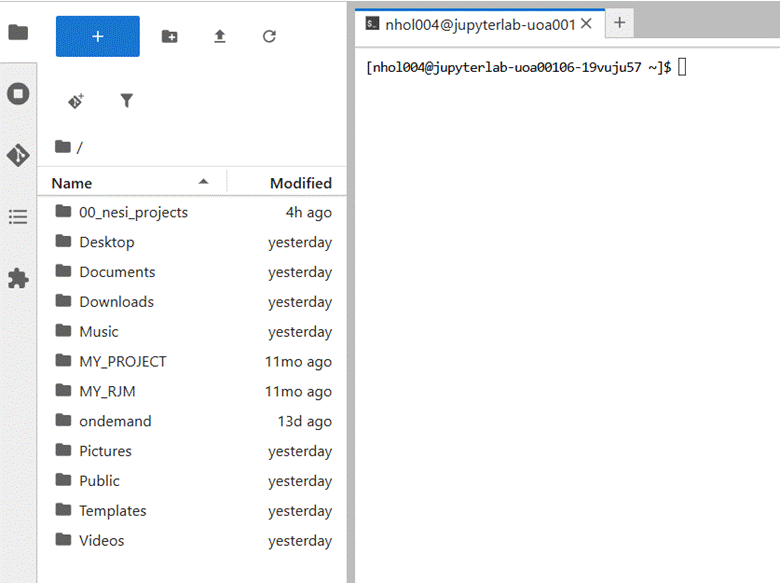
It is a matter of personal
preference which app you use to manage your files and give commands to the
operating system.
How
To Retrieve Results From A Failed Job
Jobs may fail to retrieve results if the connection used
to submit the job has stopped or if the job times out
on HPC3.
Connection Used To Submit The Job Has Stopped
After restarting the connection, if you can still see
the job results folder (model name with a _cmd1.reg suffix) there is a file in
the job results folder containing information about the job that failed to
return results. If you run nmgog again with exactly the same
model it may be possible to retrieve the results and nmgog should finish
normally by showing you the usual summary of the NONMEM job and parameter estimates.
Job Times Out On HPC3.
If the job times out then it
is possible to download to download files from HPC3
and use them to extract the latest parameter estimates. This requires several
steps which are shown here using the model “mymodel” submitted
with a control stream “mymodel.ctl”:
1.
Open a NeSI app session using
VS Code or Jupyter Lab
2.
Open the mymodel* folder. The folder name will start with the model name followed by a string showing the time the job was
submitted which is represented by the “*”.
3.
Find the files INTER, OUTPUT
and mymodel_cmd1.lst
4.
Right click on each file
in turn and download the file to the mymodel_cmd1.reg run folder on your
computer.
5.
Use the Windows File Explorer
to locate the mymodel.ctl file and the run folder mymodel_cmd1.reg.
6.
Look in mymodel_cmd1.reg and
confirm that the INTER, OUTPUT and mymodel_cmd1.lst files are in that folder. Remove
any extension such as “.txt” that may have been added to the INTER and OUTPUT files.
7.
Return to the folder containing
mymodel_cmd1.reg.
8.
Rename mymodel.ctl
to mymodel_cmd1.ctl
9.
Run the WFN nmctl command with
the “i” option. Using the “i"
option is very important. If you forget then the nmctl command will “hang”. Use
ctrl-C to stop the nmctl command then try again with the “i"
option.
nmctl mymodel_cmd1 i
10. Use the WFN nmrunmv command to rename the mymodel_cmd1
control stream and folder
nmrunmv mymodel_cmd1
mymodel
11.
The mymodel.ctl
file should now have the parameter initial estimates updated with the latest
results from the NeSI run.